Chapter 0. Introduction
0. Learning objectives
Upon completion of this chapter, the student will understand
- The motivation of enhanced sampling methods.
- Key terminologies such as phase/configuration space, ensemble, Boltzmann distribution, collective variable, free energy profile, and potential of mean force.
- The derivation of the Boltzmann distribution, potential of mean force, and free energy profile.
1. Motivation of enhanced sampling methods
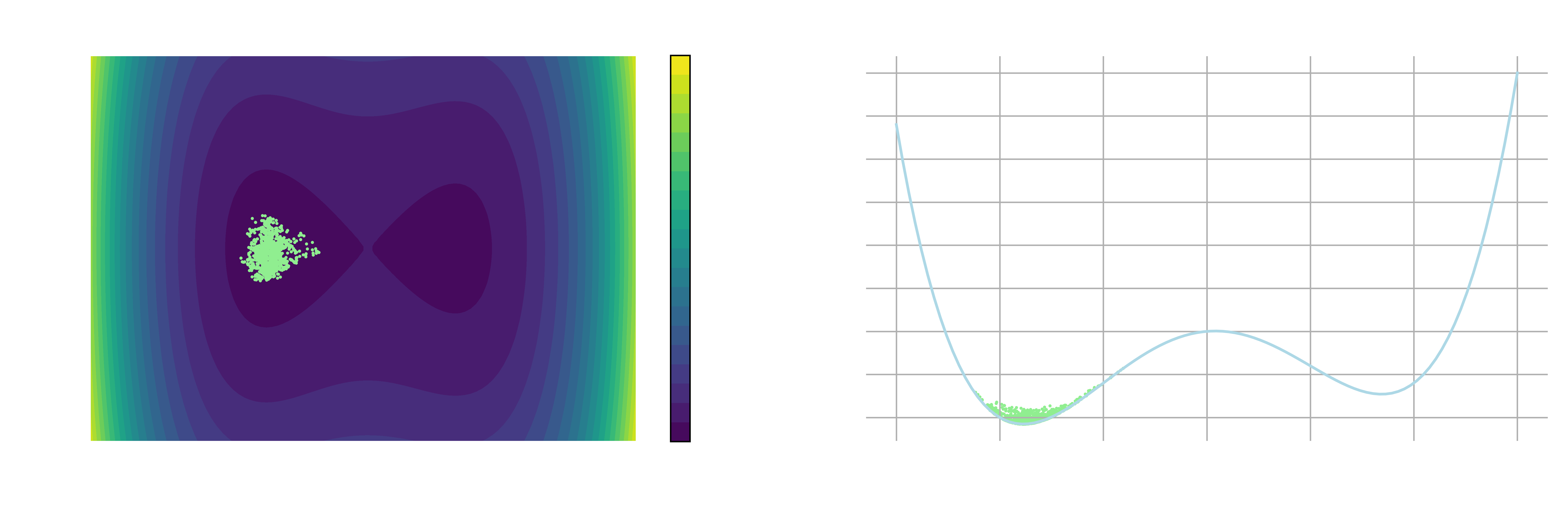
As supercomputing resources have become increasingly available in recent years1, molecular dynamics (MD) has become a standard tool for studying atomistic-level behaviors of molecular systems in various scientific disciplines2. However, an MD simulation can accurately describe the thermodynamic and structural properties of a system only if it achieves ergodicity during its simulation timescale. This means that the simulation must explore the phase space comprehensively, or equivalently, ensure that all possible configurations are sampled. This is usually a challenging task in real-world systems, as the metastable states of interest are frequently separated by substantial free energy barriers that are difficult to overcome by random fluctuations within a reasonable amount of time (see Figure 1 below, or the leftmost panel of Figure 1 in Chapter 2).
To overcome this limitation, researchers have developed various enhanced sampling methods in recent decades. One subset of these methods involves sampling along a set of predefined collective variables (CVs). These CVs, as we denote as a
2. Terminologies
Before diving into the following chapters, we will review some key concepts in statistical mechanics by introducing a few terminologies one may come across in enhanced sampling methods. This section serves as a quick review of the essential concepts to better understand the tutorials in later chapters. Readers are encouraged to visit Section 2.2 (Glossary of essential notions) in the review by Hénin et al.3 for a more exhaustive list of terms.
Timescale
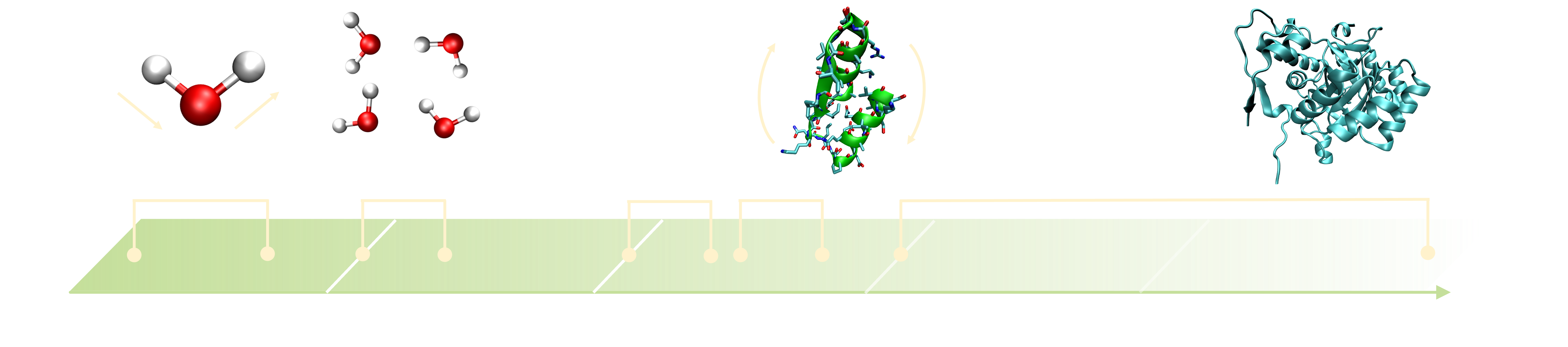
The real timescale of a molecular system can vary significantly depending on the size and complexity of the system, as well as the process of interest. As illustrated in Figure 2, the timescales of molecular motions can range from femtoseconds to seconds or even longer. For instance, bond vibrations, the fastest motion in a molecular system, typically occur on the order of femtoseconds. In contrast, biological processes such as allosteric transitions or protein-ligand binding have timescales considerably longer than bond vibrations, making the transitions between metastable states of interest rare events. This long timescale is due to the high free energy barriers separating metastable states, as discussed in the previous section.
Importantly, to ensure that the motion of atoms in a system is captured and no short-lived events are missed, such as bond vibrations and molecular rotations, the time step used in molecular dynamics simulations must be smaller than the shortest timescale in the system. This inevitably small time step makes simulations that study biological processes computationally expensive, easily requiring
Phase space and configuration space
In statistical mechanics, the phase space of a system is a space where all the possible states of the system can be represented. In three-dimensional Cartesian coordinate systems, a state is characterized by the coordinates
In contrast, the configuration space is defined by all the possible configurations (defined solely by
Microstate and macrostate
In the case where a system is composed of a larger number of particles, such as atoms or molecules, a microstate is a particular spatial arrangement of the particles in the system, i.e., a configuration. On the other hand, a macrostate, which can be defined by macroscopic thermodynamics variables (e.g., temperature, pressure, or total energy), is a collection of many possible microstates. For example, in a system composed of particles A and B that can either have an energy of 0 or 1, there are only three possible macrostates, which correspond to total energies of 0, 1, and 2, respectively. In the macrostate having a total energy of 2, there are two possible microstates: particles A and B having energies of 0 and 1, and the other way around.
Boltzmann distribution
To accurately calculate thermodynamic properties from molecular dynamics, we need to sample the Boltzmann distribution so that the system is at equilibrium. The Boltzmann distribution, sometimes called a Gibbs distribution, is a probability distribution that characterizes the distribution of states of a system in thermal equilibrium at a given temperature. It describes the probability of finding a particle in a particular energy state given its energy and the temperature of the system. Specifically, in the case where the energy levels are discrete, the normalized probability
In a phase space, where a state is defined by its continuous variables, coordinates
From Equation
Supplementary note: Maxwell-Boltzmann distribution
Different from the Boltzmann distribution, the Maxwell-Boltzmann distribution is a special case of the Boltzmann distribution. It is a probability distribution that describes the speed (
Supplementary note: Derivation of the Boltzmann distribution
The derivation of the Boltzmann distribution (Equation
- The sum of the probabilities should be 1, i.e.,
- The average energy of the particles should be
To solve for
Specifically, we have
Ensemble
In statistical mechanics, an ensemble is a collection of hypothetical copies of a system that all share the same macroscopic properties (such as temperature, pressure, volume, and chemical potential) but may differ in their microscopic details (such as the positions and velocities of individual particles). Some of the most common thermodynamic ensembles include
- NVT ensemble: The NVT ensemble is also called the canonical ensemble, where the number of particles (
- NPT ensemble: The NPT ensemble is also called the isothermal-isobaric ensemble, or the Gibbs ensemble, where the number of particles (
- NVE ensemble: The NVE ensemble is also called the microcanonical ensemble. The NVE ensemble is defined by assigning an equal probability to every microstate whose energy falls within a range centered at
Notably, given the copies of the system in an ensemble, we can calculate the ensemble average of an observable
Ergodicity
The dynamics of a system is said to be ergodic if the ensemble average of the observable
Metastable state
Metastable states are the states having a higher potential energy than the ground state but are stable enough for the system to reside for an extended (but finite) amount of time. For example, there are two metastable states in Figure 1. One has values of
Collective variable (CV)
Here, we refer to collective variables (CVs) as a
Ideally, an optimal set of CVs should be able to capture the slowest dynamics of the system and distinguish the metastable states of interest. For example, in Figure 1,
Free energy
In the canonical (NVT) ensemble, Helmholtz free energy is defined as
Free energy profile
A free energy profile is meaningful only if there is a collective variable
Supplementary note: An example of analytical free energy profile
In this example, given the potential energy
Free energy estimator
A free energy estimator is an expression or algorithm that is used to estimate free energy differences or free energy surfaces from simulation data, such as configurations, energies, and forces. Other than estimating free energy by directly measuring the probability ratios or from transition matrices, common free energy estimators include weighted histogram analysis method (WHAM)7, thermodynamic integration (TI)8, Bennett acceptance ratio (BAR)9, and multistate Bennett acceptance ratio (MBAR)10. These free energy estimators will be briefly introduced in the following chapters.
Potential of mean force (PMF)
While the term potential of mean force (PMF) is frequently used interchangeably with free energy surface, the historical definition of PMF is defined between two particles 1 and 2 and represents the effective potential energy that particle 1 experiences due to the presence of particle 2, when the positions of all other particles in the system are averaged over. It is defined based on the radial distribution function
Supplementary note: Relationship between the potential of mean force and distribution/correlation functions
1. General distribution functionsHere we consider a canonical (NVT) ensemble characterized by
2. General correlation functions
A general correlation function can be defined in terms of the distribution function
Finally, here we relate the pair correlation function/radial distribution function
3. Exercises
👨💻 Exercise 1
Given the functional form of the free energy surface in Figure 1 (
Click here for the solution.
First, we need to figure out the height of the free energy barrier going from left to right metastable states. This requires determining the local minima and maxima of the free energy surface by equating the partial derivative of
👨💻 Exercise 2
A one-dimensional simple harmonic potential with a force constant
Click here for the solution.
The one-dimensional harmonic potential is given by
Padua, D. Encyclopedia of Parallel Computing; Springer Science & Business Media, 2011. ↩︎
Gelpi, J.; Hospital, A.; Goñi, R.; Orozco, M. Molecular Dynamics Simulations: Advances and Applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. ↩︎
HéninJ.; LelièvreT.; Shirts, M. R.; Valsson, O.; Delemotte, L. Enhanced Sampling Methods for Molecular Dynamics Simulations. Living J. Comput. Mol. Sci. 2022, 4 (1). ↩︎
Hsu, W.-T.; Piomponi, V.; Merz, P. T.; Bussi, G.; Shirts, M. R. Alchemical Metadynamics: Adding Alchemical Variables to Metadynamics to Enhance Sampling in Free Energy Calculations. Journal of Chemical Theory and Computation 2023. ↩︎
Ribeiro, J. M. L.; Bravo, P.; Wang, Y.; Tiwary, P. Reweighted Autoencoded Variational Bayes for Enhanced Sampling (RAVE). The Journal of Chemical Physics 2018, 149 (7), 072301. ↩︎
Tiwary, P.; Berne, B. J. Spectral Gap Optimization of Order Parameters for Sampling Complex Molecular Systems. Proceedings of the National Academy of Sciences 2016, 113 (11), 2839–2844. ↩︎
Kumar, S.; Rosenberg, J. M.; Bouzida, D.; Swendsen, R. H.; Kollman, P. A. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I. The Method. J. Comput. Chem. 1992, 13 (8), 1011–1021. ↩︎
Kirkwood, J. G. Statistical Mechanics of Fluid Mixtures. J. Chem. Phys. 1935, 3 (5), 300–313. ↩︎
Bennett, C. H. Efficient Estimation of Free Energy Differences from Monte Carlo Data. J. Comput. Phys. 1976, 22 (2), 245–268. ↩︎
Shirts, M. R.; Chodera, J. D. Statistically Optimal Analysis of Samples from Multiple Equilibrium States. J. Chem. Phys. 2008, 129 (12), 124105. ↩︎